Abstract
Background
The proliferation and differentiation of stem cells into Germ-Like Cells (GLCs) is mediated by several growth factors and specific genes, of which some are related to long non-coding RNAs (lncRNAs). We have developed a modified differentiation process and identified a panel of GermlncRNAs related to GLCs.
Methods
Human Wharton Jelly Mesenchymal Stem Cells were treated with 25 ng/ml Bone Morphogenetic Protein (BMP)-4 and 10− 5 M all-trans retinoic acid to differentiate them into germ-like cells. To confirm the differentiation, changes in the expression of Oct-4, C-kit, Stella, and Vasa genes were assessed using quantitative Real-Time PCR (qPCR) and immunocytochemistry. QPCR was also used before and after differentiation to evaluate the changes in a lncRNA panel, using a 96-well array. Statistical analysis of the data was performed by SPSS 21.
Results
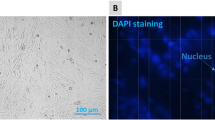
After 21 days of induction, the HWJ-MSCs derived germ-like cells were formed. Also, qPCR and immunocytochemistry showed that the pluripotent Oct4 marker was expressed in the undifferentiated HWJ-MSCs, but its expression gradually decreased in the differentiated cells. C-kit was expressed on days 7, 14, and 21 of differentiation. Both GLC markers of Stella and Vasa genes/proteins were present only in differentiated cells. Of the 44 lncRNA genes array, 36 of them showed an increase and eight genes showed a decrease.
Conclusion
Our study showed that BMP4 and RA are effective in inducing HWJ-MSCs differentiation into GLCs. In addition, our study for the first time showed changes in the lncRNAs expression during the differentiation of HWJ-MSCs into GLCs by using BMP4 and RA.








Similar content being viewed by others
References
Adamson GD, de Mouzon J, Lancaster P, Nygren K-G, Sullivan E, Zegers-Hochschild F et al (2006) World collaborative report on in vitro fertilization, 2000. Fertil Steril 85(6):1586–1622
Hou J, Yang S, Yang H, Liu Y, Liu Y, Hai Y et al (2014) Generation of male differentiated germ cells from various types of stem cells. Reproduction 147(6):R179–R88
Anawalt BD (2013) Approach to male infertility and induction of spermatogenesis. J Clin Endocrinol Metab 98(9):3532–3542
Abrao MS, Muzii L, Marana R (2013) Anatomical causes of female infertility and their management. Int J Gynecol Obstet 123:S18–S24
Sharpe RM (2010) Environmental/lifestyle effects on spermatogenesis. Philos Trans R Soc B 365(1546):1697–1712
Anderson JE, Farr SL, Jamieson DJ, Warner L, Macaluso M (2009) Infertility services reported by men in the United States: national survey data. Fertil Steril 91(6):2466–2470
Morena AR, Boitani C, Pesce M, De Felici M, Stefanini M (1996) Isolation of highly purified type A spermatogonia from prepubertal rat testis. J Androl 17(6):708–717
Leatherman J (2013) Stem cells supporting other stem cells. Front Genet 4:257
Volarevic V, Bojic S, Nurkovic J, Volarevic A, Ljujic B, Arsenijevic N et al (2014) Stem cells as new agents for the treatment of infertility: current and future perspectives and challenges. BioMed Res Int 2014
Chao KC, Chao KF, Fu YS, Liu SH (2008) Islet-like clusters derived from mesenchymal stem cells in Wharton’s Jelly of the human umbilical cord for transplantation to control type 1 diabetes. PLoS ONE 3(1):e1451
Ayatollahi M, Talaei-Khozani T, Razmkhah M (2016) Growth suppression effect of human mesenchymal stem cells from bone marrow, adipose tissue, and Wharton’s Jelly of umbilical cord on PBMCs. Iran J Basic Med Sci 19(2):145–153
Zhang H, Zheng W, Shen Y, Adhikari D, Ueno H, Liu K (2012) Experimental evidence showing that no mitotically active female germline progenitors exist in postnatal mouse ovaries. Proc Natl Acad Sci 109(31):12580–12585
Phillips BT, Gassei K, Orwig KE (2010) Spermatogonial stem cell regulation and spermatogenesis. Philos Trans R Soc B 365(1546):1663–1678
Ganesan G, Rao SM (2008) A novel noncoding RNA processed by Drosha is restricted to nucleus in mouse. RNA 14(7):1399–1410
Trovero MF, Rodríguez-Casuriaga R, Romeo C, Santiñaque FF, François M, Folle GA et al (2020) Revealing stage-specific expression patterns of long noncoding RNAs along mouse spermatogenesis. RNA Biol 17(3):350–365
Liu L, Fang F (2022) Long noncoding RNA mediated regulation in human embryogenesis, pluripotency, and reproduction. Stem Cells Int 2022:8051717
Lee T-L, Xiao A, Rennert OM (2012) Identification of novel long noncoding RNA transcripts in male germ cells. Germline development. Springer, New York, pp 105–114
Lee TL, Pang ALY, Rennert OM, Chan WY (2009) Genomic landscape of developing male germ cells. Birth Defects Res Part C 87(1):43–63
Chen Q, Meng X, Liao Q, Chen M (2019) Versatile interactions and bioinformatics analysis of noncoding RNAs. Brief Bioinform 20(5):1781–1794
Wang KC, Chang HY (2011) Molecular mechanisms of long noncoding RNAs. Mol Cell 43(6):904–914
Latifpour M, Shakiba Y, Amidi F, Mazaheri Z, Sobhani A (2014) Differentiation of human umbilical cord matrix-derived mesenchymal stem cells into germ-like cells. Avicenna J Med Biotechnol 6(4):218
Yan G, Fan Y, Li P, Zhang Y, Wang F (2015) Ectopic expression of DAZL gene in goat bone marrow-derived mesenchymal stem cells enhances the trans‐differentiation to putative germ cells compared to the exogenous treatment of retinoic acid or bone morphogenetic protein 4 signalling molecules. Cell Biol Int 39(1):74–83
Hua J, Pan S, Yang C, Dong W, Dou Z, Sidhu KS (2009) Derivation of male germ cell-like lineage from human fetal bone marrow stem cells. Reprod Biomed Online 19(1):99–105
Jafarpour Z, Soleimani M, Hosseinkhani S (2018) Efficient production of hepatocyte-like cells from human-induced pluripotent stem cells by optimizing growth factors. Int J Organ Transpl Med 9(2):77
Jaafarpour Z, Soleimani M, Hosseinkhani S, Karimi MH, Yaghmaei P, Mobarra N et al (2016) Differentiation of definitive endoderm from human induced pluripotent stem cells on hMSCs feeder in a defined medium. Avicenna J Med Biotechnol 8(1):2
Amidi F, Hoseini MA, Nia KN, Habibi M, Kajbafzadeh AM, Mazaheri Z et al (2015) Male germ-like cell differentiation potential of human umbilical cord Wharton’s Jelly-derived mesenchymal stem cells in co-culture with human placenta cells in presence of BMP4 and retinoic acid. Iran J Basic Med Sci 18(4):325
Afsartala Z, Rezvanfar MA, Hodjat M, Tanha S, Assadollahi V, Bijangi K et al (2016) Amniotic membrane mesenchymal stem cells can differentiate into germ cells in vitro. Vitro Cell Dev Biol 52(10):1060–1071
Hussain SR, Naqvi H, Ahmed F, Babu SG, Bansal C, Mahdi F (2012) Identification of the c-kit gene mutations in biopsy tissues of mammary gland carcinoma tumor. J Egypt Natl Cancer Inst 24(2):97–103
Ghasemzadeh-Hasankolaei M, Eslaminejad MB, Batavani R, Ghasemzadeh-Hasankolaei M (2015) Male and female rat bone marrow-derived mesenchymal stem cells are different in terms of the expression of germ cell specific genes. Anat Sci Int 90(3):187–196
Qin Q, Liu J, Ma Y, Wang Y, Zhang F, Gao S et al (2017) Aberrant expressions of stem cell factor/c-KIT in rat testis with varicocele. J Formos Med Assoc 116(7):542–548
Huang P, Lin LM, Wu XY, Tang QL, Feng XY, Lin GY et al (2010) Differentiation of human umbilical cord Wharton’s Jelly-derived mesenchymal stem cells into germ‐like cells in vitro. J Cell Biochem 109(4):747–754
Lacham-Kaplan O (2004) In vivo and in vitro differentiation of male germ cells in the mouse. Reproduction 128(2):147–152
Kumar K, Das K, Madhusoodan A, Kumar A, Singh P, Mondal T et al (2018) Rat bone marrow derived mesenchymal stem cells differentiate to germ cell like cells. bioRxiv:418962
Beltran M, Puig I, Peña C, García JM, Álvarez AB, Peña R et al (2008) A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial–mesenchymal transition. Genes Dev 22(6):756–769
Cacheux V, Dastot-Le Moal F, Kääriäinen H, Bondurand N, Rintala R, Boissier B et al (2001) Loss-of-function mutations in SIP1 Smad interacting protein 1 result in a syndromic Hirschsprung disease. Hum Mol Genet 10(14):1503–1510
Yu M, Tian T, Zhang J, Hu T (2022) miR-141-3p protects against blood–brain barrier disruption and brain injury after intracerebral hemorrhage by targeting ZEB2. J Clin Neurosci 99:253–260
Lou G, Dong X, Xia C, Ye B, Yan Q, Wu S et al (2016) Direct targeting sperm-associated antigen 9 by miR-141 influences hepatocellular carcinoma cell growth and metastasis via JNK pathway. J Exp Clin Cancer Res 35(1):1–10
Yadav RP, Kotaja N (2014) Small RNAs in spermatogenesis. Mol Cell Endocrinol 382(1):498–508
Kotaja N (2014) MicroRNAs and spermatogenesis. Fertil Steril 101(6):1552–1562
Abu-Halima M, Hammadeh M, Schmitt J, Leidinger P, Keller A, Meese E et al (2013) Altered microRNA expression profiles of human spermatozoa in patients with different spermatogenic impairments. Fertil Steril 99(5):1249–1255e16
Su Y, Zhou LL, Zhang YQ, Ni LY (2019) Long noncoding RNA HOTTIP is associated with male infertility and promotes testicular embryonal carcinoma cell proliferation. Mol Genet Genom Med 7(9):e870
Hadziselimovic F, Gegenschatz-Schmid K, Verkauskas G, Demougin P, Bilius V, Dasevicius D et al (2017) GnRHa treatment of cryptorchid boys affects genes involved in hormonal control of the HPG axis and fertility. Sex Dev 11(3):126–136
Flanagan CA, Manilall A (2017) Gonadotropin-releasing hormone (GnRH) receptor structure and GnRH binding. Front Endocrinol 8:274
Lin J, Mao J, Wang X, Ma W, Hao M, Wu X (2019) Optimal treatment for spermatogenesis in male patients with hypogonadotropic hypogonadism. Medicine 98:31
Nishida H, Miyagawa S, Vieux-Rochas M, Morini M, Ogino Y, Suzuki K et al (2008) Positive regulation of steroidogenic acute regulatory protein gene expression through the interaction between Dlx and GATA-4 for testicular steroidogenesis. Endocrinology 149(5):2090–2097
Hadziselimovic F, Verkauskas G, Vincel B, Stadler MB (2019) Testicular expression of long non-coding RNAs is affected by curative GnRHa treatment of cryptorchidism. Basic Clin Androl 29:18
Vigneau S, Rohrlich P-S, Brahic M, Bureau J-F (2003) Tmevpg1, a candidate gene for the control of Theiler’s virus persistence, could be implicated in the regulation of gamma interferon. J Virol 77(10):5632–5638
Collier SP, Collins PL, Williams CL, Boothby MR, Aune TM (2012) Cutting edge: influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. J Immunol 189(5):2084–2088
Salviano-Silva A, Lobo-Alves SC, Almeida RC, Malheiros D, Petzl-Erler ML (2018) Besides pathology: long non-coding RNA in cell and tissue homeostasis. Noncoding RNA 4(1)
Klein B, Haggeney T, Fietz D, Indumathy S, Loveland KL, Hedger M et al (2016) Specific immune cell and cytokine characteristics of human testicular germ cell neoplasia. Hum Reprod 31(10):2192–2202
Gu J, Wang Y, Wang X, Zhou D, Shao C, Zhou M et al (2018) Downregulation of lncRNA GAS5 confers tamoxifen resistance by activating miR-222 in breast cancer. Cancer Lett 434:1–10
Li M, Xie Z, Wang P, Li J, Liu W, Liu Z et al (2018) The long noncoding RNA GAS5 negatively regulates the adipogenic differentiation of MSCs by modulating the miR-18a/CTGF axis as a ceRNA. Cell Death Dis 9(5):1–13
Yacqub-Usman K, Pickard MR, Williams GT (2015) Reciprocal regulation of GAS5 lncRNA levels and mTOR inhibitor action in prostate cancer cells. Prostate 75(7):693–705
Wang J, Gong X, Tian GG, Hou C, Zhu X, Pei X et al (2018) Long noncoding RNA growth arrest-specific 5 promotes proliferation and survival of female germline stem cells in vitro. Gene 653:14–21
Lanz RB, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY et al (1999) A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell 97(1):17–27
Yan R, Wang K, Peng R, Wang S, Cao J, Wang P et al (2016) Genetic variants in lncRNA SRA and risk of breast cancer. Oncotarget 7(16):22486
Hube F, Velasco G, Rollin Jm, Furling D, Francastel C (2011) Steroid receptor RNA activator protein binds to and counteracts SRA RNA-mediated activation of MyoD and muscle differentiation. Nucleic Acids Res 39(2):513–525
Xu B, Yang W-H, Gerin I, Hu C-D, Hammer GD, Koenig RJ (2009) Dax-1 and steroid receptor RNA activator (SRA) function as transcriptional coactivators for steroidogenic factor 1 in steroidogenesis. Mol Cell Biol 29(7):1719–1734
Luo P, Jing W, Zhu M, Li N-D, Zhou H, Yu M-X et al (2017) Decreased expression of LncRNA SRA1 in hepatocellular carcinoma and its clinical significance. Cancer Biomark 18(3):285–290
Acknowledgements
Thanks to Dr. Mostafa Latifpour and his colleagues to provide the Human Wharton Jelly Mesenchymal Stem Cells as a gift.
Funding
This study was derived from a thesis for an M.Sc. degree in the field of medical biotechnology (Grant or Code Number: 950214021) at Gorgan School of Advanced Technologies in Medicine, Golestan University of Medical Sciences, Gorgan, Iran.
Author information
Authors and Affiliations
Contributions
SGH: Performing experiments, acquisition of data, analysis and interpretations of data, manuscript drafting, revision of the manuscript. MSH: Contribution to study design. GAF: Contribution to data analysis and manuscript drafting. JTA: Contribution to the evaluation of experiments and manuscript drafting. MS: Contribution to study design. SR: Contribution to manuscript revision. NM: Study design and concept, participation in literature bibliography, data acquisition and analysis, manuscript drafting, and critical revision of the manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethical approval
The study was approved by the Committee of Ethics, Golestan University of Medical Sciences (Code of Ethics: IR.GOUMS.REC.1394.34).
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ghasemi, S., Shafiee, M., Ferns, G.A. et al. Differentiation of Human Wharton Jelly Mesenchymal Stem Cells into Germ-Like Cells; emphasis on evaluation of Germ-long non-coding RNAs. Mol Biol Rep 49, 11901–11912 (2022). https://doi.org/10.1007/s11033-022-07961-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-07961-6